3 Adjacency matrix
Adjacency-Matrix Representation
An adjacency-matrix representation of a graph is $V\times V$ matrix of Boolean values, with entry in row $v$ and column $w$ defined to be 1 if there is an edge connecting vertex $v$ and vertex $w$ in the graph, and to be 0 otherwise.
This representation is well suited for dense graphs.
For processing all of the vertices this implementation requires (at least ) time proportional to $V$, no matter how many vertices exist.
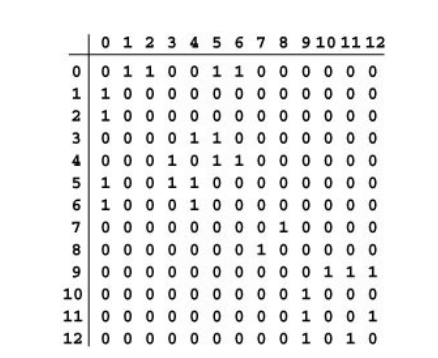
This matrix is symmetric about diagonal because of undirected graph.
Graph ADT implementation (adjacency-matrix)
class DenseGRAPH
{
int Vcnt,Ecnt; bool digraph;
vector <vector <bool> > adj;
public:
DenseGRAPH(int V, bool digraph = false):
adj(V), Vcnt(V), Ecnt(0), digraph(digraph)
{
for(int i = 0 ; i<V; i++)
adj[i].assign(V,false);
}
int V() const { return Vcnt; }
int E() const { return Ecnt; }
bool directed() const { return digraph; }
void insert(Edge e)
{
int v = e.v , w = e.w;
if(adj[v][w] == false ) Ecnt++;
adj[v][w] = true;
if(!digraph) adj[w][v] = true;
}
void remove(Edge e)
{ int v = e.v, w = e.w;
if(adj[v][w] == true) Ecnt--;
adj[v][w] = false;
if(!digraph) adj[w][v] = false;
}
bool edge(int v, int w) const
{ return adj[v][w];}
class adjIterator;
friend class adjIterator;
};
Iterator for adjacency-matrix representation
class DenseGRAPH::adjIterator
{ const DenseGRAPH &G;
int i,v;
public:
adjIterator(const DenseGRAPH &G, int v):
G(G),v(v),i(-1) {}
int beg()
{ i = -1 ; return nxt();}
int nxt()
{
for(i++; i<G.V(); i++)
if(G.adj[v][i] == true) return i;
return -1;
}
bool end()
{ return i>=G.V();}
};